© Stevenn VOLANT / Amine GHOZLANE / Javier PIZZARO CERDA / Juan Jose QUEREDA TORRES / IPL / CNRS Images
Reference
20170074_0012
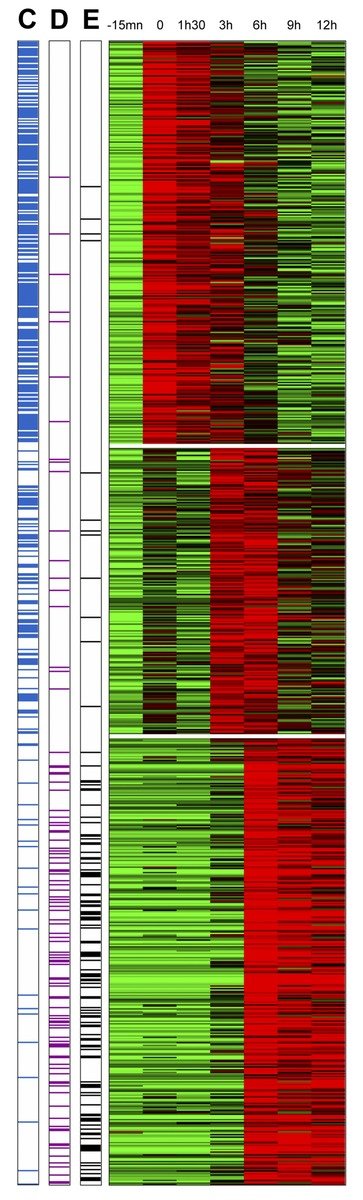
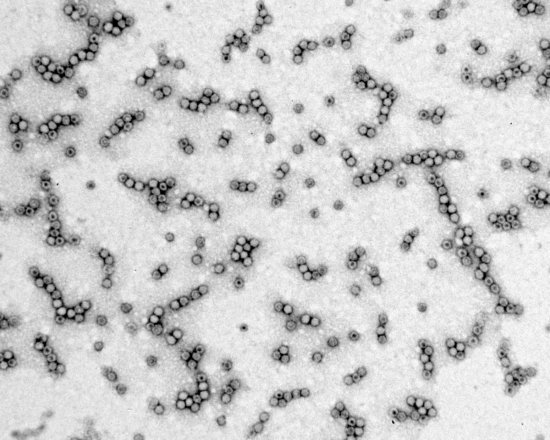
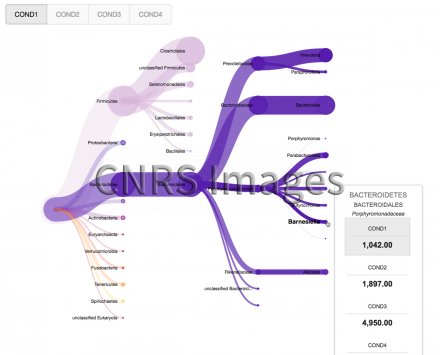
Arbre d’abondance métagénomique d'un microbiome intestinal de souris
Taxonomic tree of dynamic abundance obtained using quantitative metagenomic analysis. This analysis involves identifying associations between a microbiome, different environmental conditions and individuals with particular characteristics (diseases, geographical origin, etc.). Here, research scientists have studied the intestinal microbiome of mice infected with the bacterium Listeria monocytogenes. In this experiment, the abundance of Alloprevotella and Allobaculum bacteria populations is compared to isoleucine S (LLS) gene expression. This image was generated using the SHAMAN (SHiny Application for Metagenomic ANalysis) software package. The recent development of new sequencing technologies means these organisms can be studied at the level of the genome. For Listeria, for example, this allows the identification of new factors contributing to bacterial virulence, notably during the intestinal phase of the infection.
The use of media visible on the CNRS Images Platform can be granted on request. Any reproduction or representation is forbidden without prior authorization from CNRS Images (except for resources under Creative Commons license).
No modification of an image may be made without the prior consent of CNRS Images.
No use of an image for advertising purposes or distribution to a third party may be made without the prior agreement of CNRS Images.
For more information, please consult our general conditions