© Olivier GASCUEL / Matthieu JUNG / Martine PEETERS / C3BI / IRD / CNRS Images
Reference
20170075_0001
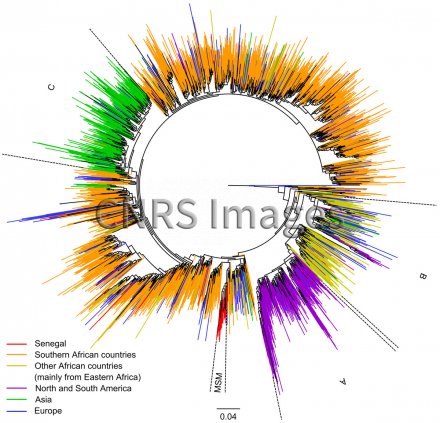
Arbre phylogénétique décrivant la propagation du sous-type C du VIH-1
Phylogenetic tree describing the propagation of HIV-1 subtype C, generated with the PhyML software package using more than 3000 sequences of the virus from across the world. The tree provides an overview of how the sequences have evolved, with the colour scheme used to link this evolution to geography and risk factors. Each branch (coloured line) represents the evolution of a viral sequence. The length of the branches varies according to the estimated number of mutations. The junctions between the branches correspond to points of inter-patient transmission or intra-patient diversification. The scale of 0.04 corresponds to a nucleotide mutation rate of 4%. As the average rate of mutation is around 0.003 mutations per nucleotide per year, this scale represents slightly more than 10 years of evolution. The letters A, B and C refer to subgroups of subtype C, uniting strains that correspond to geographically linked isolates. For example, subgroup A was principally identified in America and subgroup C chiefly in Asia. This homogeneity indicates a recent, and sometimes single, introduction, such as in the case of Asia, unlike in Africa and Europe. The red part, MSM (men who have sex with men), shows that in Senegal, the introduction of subtype C to this specific population was also the result of a single introduction. Research into HIV-1 introductions at country and continent level, and within risk groups, helps us better understand the mechanisms of epidemics and control the spread of the virus more effectively.
The use of media visible on the CNRS Images Platform can be granted on request. Any reproduction or representation is forbidden without prior authorization from CNRS Images (except for resources under Creative Commons license).
No modification of an image may be made without the prior consent of CNRS Images.
No use of an image for advertising purposes or distribution to a third party may be made without the prior agreement of CNRS Images.
For more information, please consult our general conditions